Documentation
Data and Sources
Brassicaceae
- Comparative Evolution (CoGe)
- EnsemblPlants
- http://plants.ensembl.org/Arabidopsis_thaliana/Info/Index
- http://plants.ensembl.org/Multi/Search/Results/species_plants?db=core;q=arabidopsis;species=all;collection=all;site=ensemblunit;
- http://plants.ensembl.org/Multi/Search/Results?species=all;idx=;q=NRPD1B;site=ensemblunit
- http://plants.ensembl.org/Multi/Search/Results?species=all;idx=;q=KTF1;site=ensemblunit
- Phytozome
- Salk Institute Genomic Analysis Laboratroy (SIGnAL)
- 1001 Genomes
Oryzeae
- Comparative Evolution (CoGe)
- EnsemblPlants
- GigaScience database (GigaDB)
- National Center for Biotechnology Information (NCBI) database biosample for 3000 genomes project
- OryzaSNP
- Rice Annotation Project Database (RAP-DB)
- Rice Genome Annotation Project (RGAP)
- 3k RGP and OryzaSNP data repository
Programs
AGO Hook Identification
- Self-generated python scripts
- Available on my GitHub account
- Identifies AGO Hooks within a protein sequence
- Requirements to run...
- FASTA file with sequence(s) of interest
DnaSP
- Software of analyzing DNA sequence polymorphisms (SNPs)
- Requirements to run...
- Alignment file containing sequences of interest
Genome Analysis Toolkit (GATK)
- Offer by Broad Institute
- FASTA file converter/maker
- Software for analyzing high-throughput next-gen sequencing data
- Requirements to run...
InterPro Scan
- Offered by EMBL-EBI
- Identifies known domains within a protein of interest
- Requirements to run...
- Installation of SOAPpy, PyXML, and fpconst Python Modules
- All modules downloaded and used available from python** https://pypi.python.org/pypi?%3Aaction=index
LAST
- Offered by Computational Biology Research Center (CBRC)
- Homology search tool
- Requirements to run...
- FASTA files of sequences to be aligned/searched
MEGA6
- Multitool program for bioinformatic work
- Requirements to run...
- FASTA file with sequences to be analyzed
RADAR
- Offered by EMBL-EBI
- Identifies repeating sequences within a protein of interest
- Requirements to run...
- Installation of Cython (http://cython.org) and lfasta
RAxML
- Offered by the Exelixis lab
- Phytogenetic tree building tool
- Requirements to run...
- Alignment file containing sequences of interest
Variant Tools
- Software for manipulating, selecting and annotating next-gen sequencing data
- Requirements to run...
Science
Introduction/Background
Nuclear RNA Polymerase V (NRPE) functions in miRNA directed DNA methylation pathway leading to transcriptional gene silencing. RNA Pol V arose through a whole genome duplication event and subsequently evolved a new and unique function from the ancestral RNA Pol II. The largest subunit of RNA Pol V (E1) is unique to RNA Pol V and contains RNA binding domains as well as recently discovered motifs in the C-terminal domain (CTD). Of these two motifs, one is important for protein-protein interactions, but the other motifs is still of unknown function. The first identified motif is a dipeptide motif, consisting of Glycine (G) and Tryptophan (W), known as "AGO Hooks", and important for protein-protein interactions occurring at the CTD. AGO Hooks got there name because the motif was first identified in proteins that interact ARGONAUT (AGO) proteins. The other motif present is more variable in length as well as conservatrion but are always found spread thoughout the CTD. Previous studies in the plant family Brassicaceae reveal a great deal of variation in the number of repeats present between species of a family as well as indicate that the repeated sequence is unique to the family. Several other well studied plant families also ?possess a repeat sequence unique to that particular family. It is hypothesized that these repeats have an unknown function and have arisen through mismatching during homologous recombination, allowing for the repeats to expand or contract. In addition to investigating NRPE1 evolution, one of its interactiving proteins, SPT5L, will be analyzed as well.
Results
Nuclear RNA Polymerase Subunit 1 (NRPE1)
Arabidopsis thaliana
Fig.1. NRPE1 coding sequence and genomic sequences aligned for visualization of exons. Annotation included for conserved domains (pink), AGO Hooks (red), and unique repeat (gray).
Fig. 2. NRPE1 Carboxy terminal domain (CTD) tandemly repeating sequence of unknown function.
Analysis also done for twelve (12) other species within Brassicaceae:
A. lyrata, B. rapa, B. oleraceae, N. paniculata, C. rubella, C. sativa, S. irio, L. alabamica, E. parvula, E. salsugineum, A. arabicum, and T. hassleriana
SNP Analysis
Using A. thaliana SNP information to investigate selection on this gene, Tajima's D was determined and reported as -2.117 for the protein overall.
First 100 accessions were analyzed to get an idea of the amount of selection on this gene as well as computation time needed.
Fig. 3. Tajima's D analysis of 100 accessions. Sliding window of 100 nt.
Fig. 4. Tajima's D analysis of 100 accessions viewed with 20 nt sliding window.
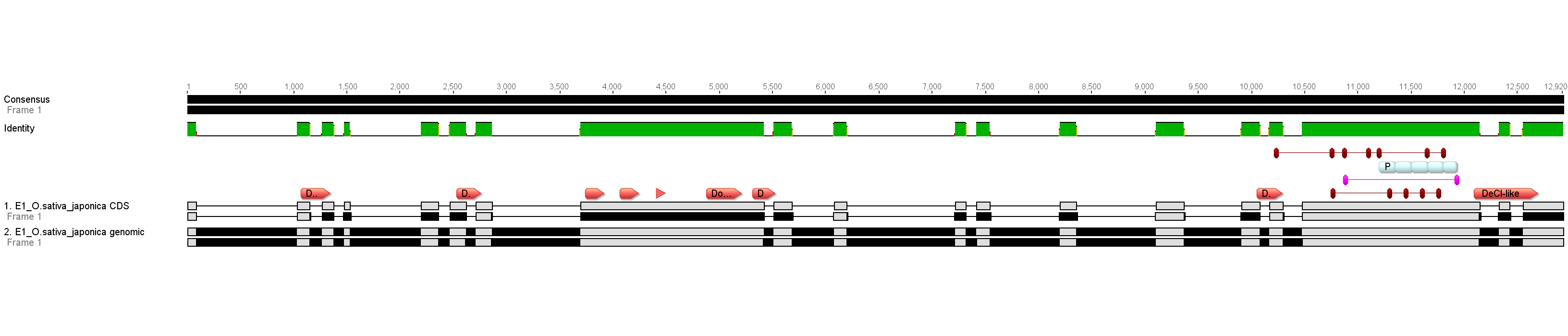
Oryza sativa ssp japonica
Fig.1. NRPE1 coding sequence and genomic sequences aligned for visualization of exons. Annotation included for conserved domains (pink), AGO Hooks (red), and unique repeat (gray).
Fig. 2. NRPE1 Carboxy terminal domain (CTD) tandemly repeating sequence of unknown function.
Analysis also done for eleven (10) other Oryza species:
O. sativa ssp indica, O.barthii, O.brachyantha, O. glaberrima, O. glumaepatula, O. longistaminata, O. meridionalis, O. nirvara, O. punctata, and O. rufipogon.
Transcription elongation factor Suppressor of TY 5 like (SPT5L)
Arabidopsis thaliana
Fig.1 STP5L coding sequence and genomic sequences aligned for visualization of exons. Annotation included for conserved domains (pink), AGO Hooks (red), and unique repeat (gray).
Fig. 2 One of the two tandemly repeating sequence of unknown function found within the Carboxy terminal domain (CTD).
Fig. 3 The second tandemly repeating sequence of unknown function found within the Carboxy terminal domain (CTD).
Only A. thaliana, A. lyrata, and B. rapa have been analyzed at this point.
Oryza sativa
Coming soon!
Methods
NRPE1 and SPT5L genomic sequences were identified and obtained by homology search using each gene sequence from Arabidopsis thaliana and Oryza sativa spp japonica for the Brassicaceae and Oryzeae families, respectfully . Refer to Data and Sources for databases used to . FGENESH+ (see above) was used to identify the coding sequence (CDS) for each gene if the CDS was not accompaning the genomic in the databases. Gene sequences for each species in a family were then annotated with Geneious using A. thaliana for Brassicaceae and O. sativa ssp japonica for Oryzeae. as done previously. Single Nucleotide Polymorphism data
Discussion
References
Access, O. (2014). The 3,000 rice genomes project. GigaScience, 3, 7. doi:10.1186/2047-217X-3-7
Matzke, M. a, & Mosher, R. a. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature Reviews. Genetics, 15(6), 394--408. doi:10.1038/nrg3683
Nelson, A. D. L., Forsythe, E. S., Gan, X., Tsiantis, M., & Beilstein, M. a. (2014). Extending the model of Arabidopsis telomere length and composition across Brassicaceae. Chromosome Research?: An International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology, 22(2), 153--66. doi:10.1007/s10577-014-9423-y
Kane, J., Freeling, M., & Lyons, E. (2010). The evolution of a high copy gene array in arabidopsis. Journal of Molecular Evolution, 70(6), 531--544.