fastStructure v1.0 using Atmosphere
Rationale and background:
Tools for estimating population structure and from genetic data are now used in a wide variety of applications in population genetics. In addition, identifying the degree of admixture in individuals and inferring the population of origin of specific loci in these individuals is relevant for a variety of problems in population genetics. With decreasing costs in sequencing and genotyping technologies, there is an increasing need for fast and accurate tools to infer population structure from very large genetic data sets. However, inferring population structure and identifying the degree of admixture in large modern data sets imposes severe computational challenges. fastStructure is a fast algorithm for inferring population structure from large SNP genotype data. It is based on a variational Bayesian framework for posterior inference and is written in Python2.x (Anil Raj et al., Genetics Jan 2014). It is an efficient algorithm for approximate inference of the model underlying the STRUCTURE program.Variational methods pose the problem of computing relevant posterior distributions as an optimization problem, allowing the use of recent advances in optimization theory to develop fast inference tools. In addition, useful heuristic scores can be used to identify the number of populations represented in a dataset and a new hierarchical prior to detect weak population structure in the data. The variational algorithms are almost two orders of magnitude faster than STRUCTURE and achieve accuracies comparable to those of ADMIXTURE. Furthermore, the results show that the heuristic scores for choosing model complexity provide a reasonable range of values for the number of populations represented in the data, with minimal bias towards detecting structure when it is very weak.
This tutorial will take users through steps of:
- Launching the fastStructure Atmosphere image
- Running fastStructure on an test data
Please work through the tutorial and add your comments on the bottom of this page. Or send comments per email to support@cyverse.org. Thank you.
Learn about allocations
Learn about CyVerse's allocation policies here.
Part 1: Connect to an instance of an Atmosphere Image (Virtual Machine)
Step 1. Go to https://atmo.cyverse.org and log in with your CyVerse credentials.
Step 2. Create a new project (faststructure) and add some description after log-in.
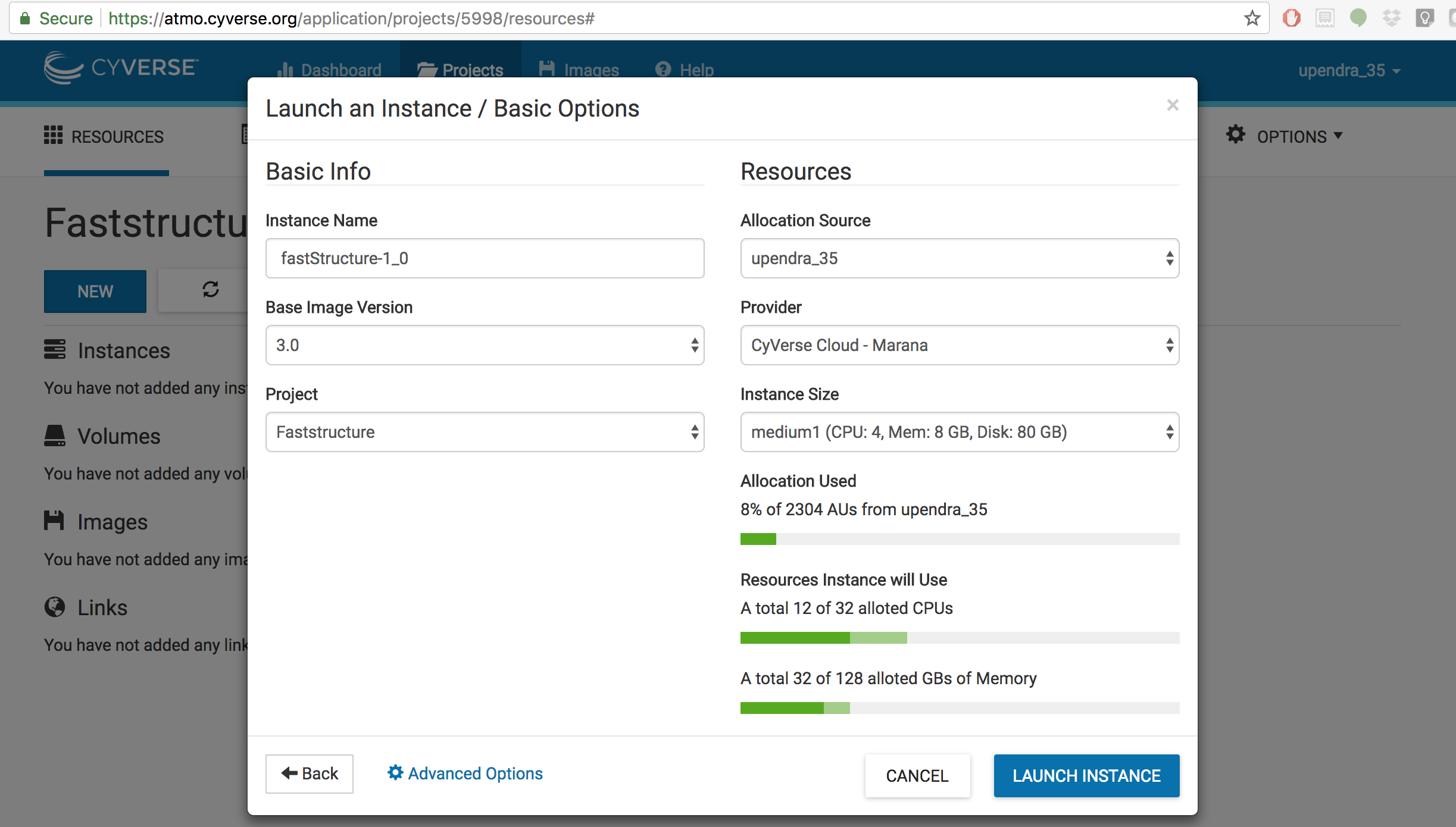
Step 3. Click on the Launch New Instance button and search for fastStructure-1.0 image.
Step 4. Click Launch Instance. It will take 2-5 minutes for the cloud instance to be launched.
Note: Instances can be configured for different amounts of CPU, memory, and storage depending on user needs. This tutorial can be accomplished with the medium1 instance size, medium1 (4 CPUs, 8 GB memory, 80 GB root)
Part 2: Set up a fastStructure run using the Terminal window
Step 1.
- Open the Terminal on mac. Add the ssh details along with your IP address to connect the instance through the terminal
$ ssh <username>@Ipaddress
- or using webshell
Step 2. Get oriented.
- You will find fastStructure software in "/opt" folder. All the dependencies for running fastStructure are located in "/opt/fastStructure" but you need to change the permissions before using it
$ ls /opt/fastStructure-1.0/ build distruct.py fastStructure.pyx LICENSE parse_bed.pyx parse_str.c parse_str.so setup.py test chooseK.py fastStructure.c fastStructure.so parse_bed.c parse_bed.so parse_str.pyx README.md structure.py vars
Step 3. The staged example data can be found in folder "fastStructure/test" within "opt" folder. List its contents with the ls command:
$ ls /opt/fastStructure-1.0/test testdata.bed testoutput_logistic.3.log testoutput_logistic.3.varP testoutput_simple.3.meanP testoutput_simple.3.varQ testdata.bim testoutput_logistic.3.meanP testoutput_logistic.3.varQ testoutput_simple.3.meanQ testdata.fam testoutput_logistic.3.meanQ testoutput_simple.3.log testoutput_simple.3.varP
The testdata.bed file (with corresponding test.fam and test.bim) contains genotypes sampled for 200 individuals at 500 SNP loci. There are also other testdata outpuut files. Let's copy the files
$ cd ~ $ cp -R /opt/fastStructure-1.0/test . $ rm test/*output* $ ls test testdata.bed testdata.bim testdata.fam
Step 4. Set up a fastStructure test run. Executing the code with the provided test data should generate a log file identical to the ones in test/, as a first check that the source code has been downloaded and compiled correctly. The algorithm scales linearly with number of samples, number of loci and value of K; the expected runtime for a new dataset can be computed from the runtime in the above log file.
$ python /opt/fastStructure-1.0/structure.py -K 2 --input=test/testdata --output=test/testoutput_simple --full --seed=100 $ python /opt/fastStructure-1.0/structure.py -K 3 --input=test/testdata --output=test/testoutput_simple --full --seed=100 $ python /opt/fastStructure-1.0/structure.py -K 4 --input=test/testdata --output=test/testoutput_simple --full --seed=100
$ ls test testdata.bed testoutput_simple.2.meanP testoutput_simple.3.log testoutput_simple.3.varQ testoutput_simple.4.varP testdata.bim testoutput_simple.2.meanQ testoutput_simple.3.meanP testoutput_simple.4.log testoutput_simple.4.varQ testdata.fam testoutput_simple.2.varP testoutput_simple.3.meanQ testoutput_simple.4.meanP testoutput_simple.2.log testoutput_simple.2.varQ testoutput_simple.3.varP testoutput_simple.4.meanQ
Step 5. Choosing the model complexity: In order to choose the appropriate number of model components that explain structure in the dataset, it is recommended to run the algorithm for multiple choices of K. FastStructure provides a utility tool to parse through the output of these runs and provide a reasonable range of values for the model complexity appropriate for this dataset.
Assuming the algorithm was run on the test dataset for choices of K ranging from 1 to 5, and the output flag was --output=test/testoutput_simple, you can obtain the model complexity by doing the following:
$ python /opt/fastStructure-1.0/chooseK.py --input=test/testoutput_simple Model complexity that maximizes marginal likelihood = 2 Model components used to explain structure in data = 2
Here the best model to explain the data is 2.
Step 6. Visualizing the admixture proportions. In order to visualize the expected admixture proportions inferred by fastStructure, fastStructure provided a simple tool to generate Distruct plots using the mean of the variational posterior distribution over admixture proportions. The samples in the plot will be grouped according to population labels inferred by fastStructure. However, if the user would like to group the samples according to some other categorical label (e.g., geographic location), these labels can be provided as a separate file using the flag --popfile. The order of labels in this file (one label per row) should match the order of samples in the input data files.
$ python /opt/fastStructure-1.0/distruct.py -K 2 --input=test/testoutput_simple --output=test/testoutput_simple_distruct_K2.pdf $ python /opt/fastStructure-1.0/distruct.py -K 3 --input=test/testoutput_simple --output=test/testoutput_simple_distruct_K3.pdf $ python /opt/fastStructure-1.0/distruct.py -K 4 --input=test/testoutput_simple --output=test/testoutput_simple_distruct_K4.pdf
$ ls testdata.bed testoutput_simple.2.varP testoutput_simple.3.varQ testoutput_simple_distruct_K2.pdf testdata.bim testoutput_simple.2.varQ testoutput_simple.4.log testoutput_simple_distruct_K3.pdf testdata.fam testoutput_simple.3.log testoutput_simple.4.meanP testoutput_simple_distruct_K4.pdf testoutput_simple.2.log testoutput_simple.3.meanP testoutput_simple.4.meanQ testoutput_simple.2.meanP testoutput_simple.3.meanQ testoutput_simple.4.varP testoutput_simple.2.meanQ testoutput_simple.3.varP testoutput_simple.4.varQ
Step 7. Downloading the files using cyberduck.
For a new dataset, it is recommend executing fastSTRUCTURE, for multiple values of K to obtain a reasonable range of values for the appropriate model complexity required to explain structure in the data, as well as ancestry estimates at those model complexities. For improved ancestry estimates and to identify subtle structure, it is recommend executing fastSTRUCTURE with the logistic prior at values of K similar to those identified when using the simple prior.