Variant annotation using SnpEff in DE
SnpEff: Genomic variant annotations and functional effect prediction toolbox.
Rationale
SnpEff is a variant annotation and effect prediction tool. It annotates and predicts the effects of variants on genes (such as amino acid changes). The inputs are predicted variants (SNPs, insertions, deletions and MNPs). The input file is usually obtained as a result of a sequencing experiment, and it is usually in variant call format (VCF). SnpEff analyzes the input variants. It annotates the variants and calculates the effects they produce on known genes (e.g. amino acid changes).
Reference
"A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3.", Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. Fly (Austin). 2012 Apr-Jun;6(2):80-92. PMID: 22728672 [PubMed - in process]
Databases
In order to produce the annotations, SnpEff requires a database. SnpEff build these databases using informations from trusted resources.
By default SnpEff downloads and install databases automatically (since version 4.0)
Currently, there are pre-built database for over 20,000 reference genomes. This means that most cases are covered. In some very rare occasions, people need to build a database for an organism not currently supported (e.g. the genome is not publicly available). In most cases, this can be done and follow the instructions in the section of How to build your own SnpEff database.
Which databases are supported? You can find out all the supported databases by clicking this file snpeff_databases.csv. The csv file consists of 2 columns, the first column denotes the Genome and the second column denotes the Organism. Here is a short preview of the file..
Genome Organism 129S1_SvImJ_v1.86 Mus_musculus_129s1svimj AKR_J_v1.86 Mus_musculus_akrj A_J_v1.86 Mus_musculus_aj Abiotrophia_defectiva_atcc_49176 Abiotrophia_defectiva_atcc_49176 Acanthamoeba_castellanii_str_neff Acanthamoeba_castellanii_str_neff Acaryochloris_marina_mbic11017 Acaryochloris_marina_mbic11017 Acetivibrio_ethanolgignens Acetivibrio_ethanolgignens Acetobacter_aceti_1023 Acetobacter_aceti_1023 Acetobacter_aceti_nbrc_14818 Acetobacter_aceti_nbrc_14818 ......................... .........................
If you want to know more details such as source data (where was the genome reference data obtained from) then please refer this snpeff_databases_detailed.csv .
GRCh37 instead of hg19, or GRCm38 instead of mm10, and so on. 2. Running SnpEff
2.1 Basic example: Annotate using SnpEff
Let's assume you have a VCF file and you want to annotate the variants in that file you can annotate the file by launching snpEff app in DE.
All files are located for Basic example is located in the Community Data directory of the CyVerse Discovery Environment at the following path:
Community Data > iplantcollaborative > example_data > snpEff (/iplant/home/shared/iplantcollaborative/example_data/snpEff)
2.1.1 After logging into Discovery Environment, click on the app window and in the search box, enter snpeff. You will see two apps - SnpEff-4.3.1 and SnpEff-build-4.3.1
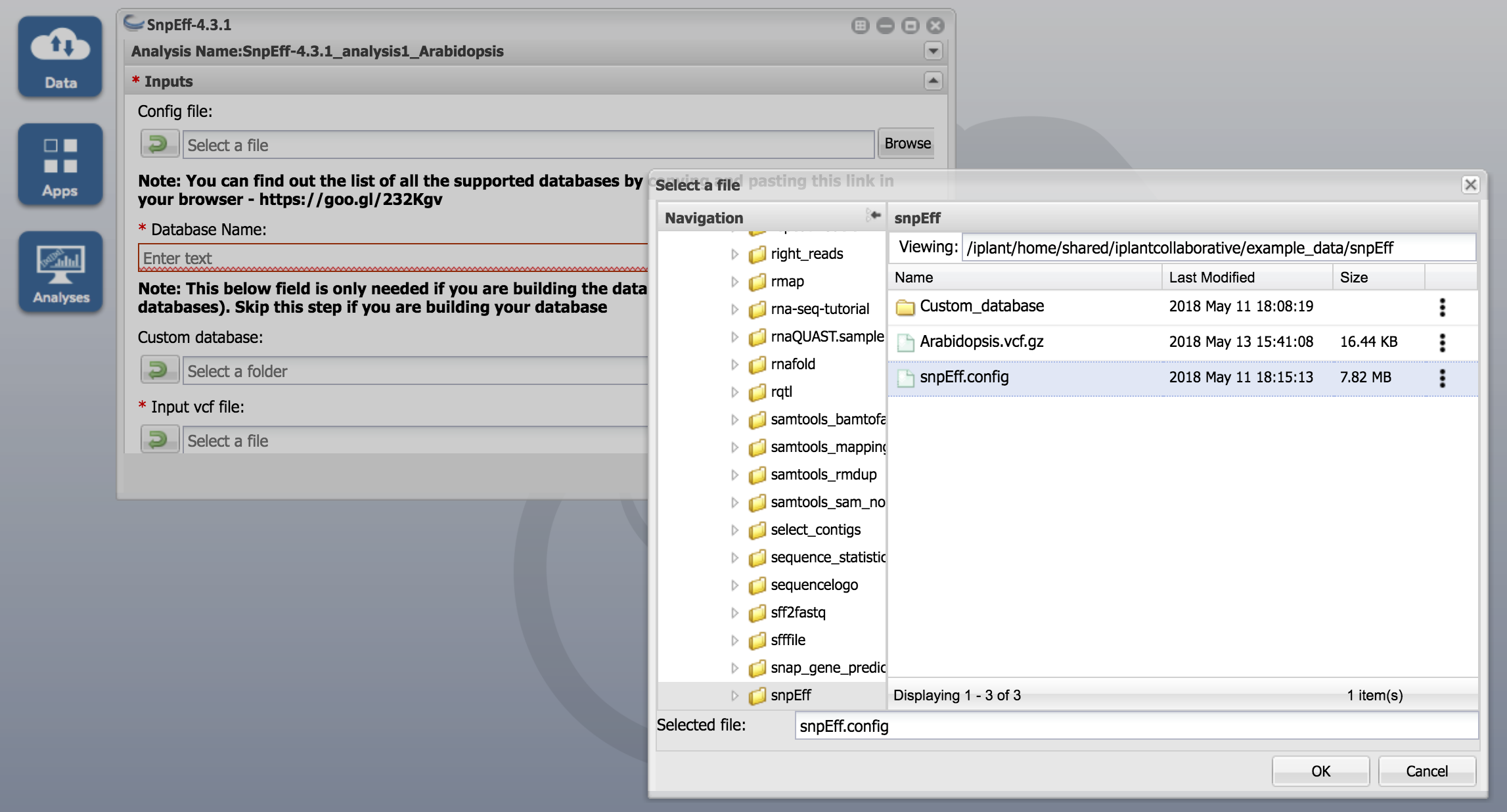
Click on the SnpEff-4.3.1 app and enter "SnpEff-4.3.1_analysis1_Arabidopis" under the Analysis Name
Inputs:
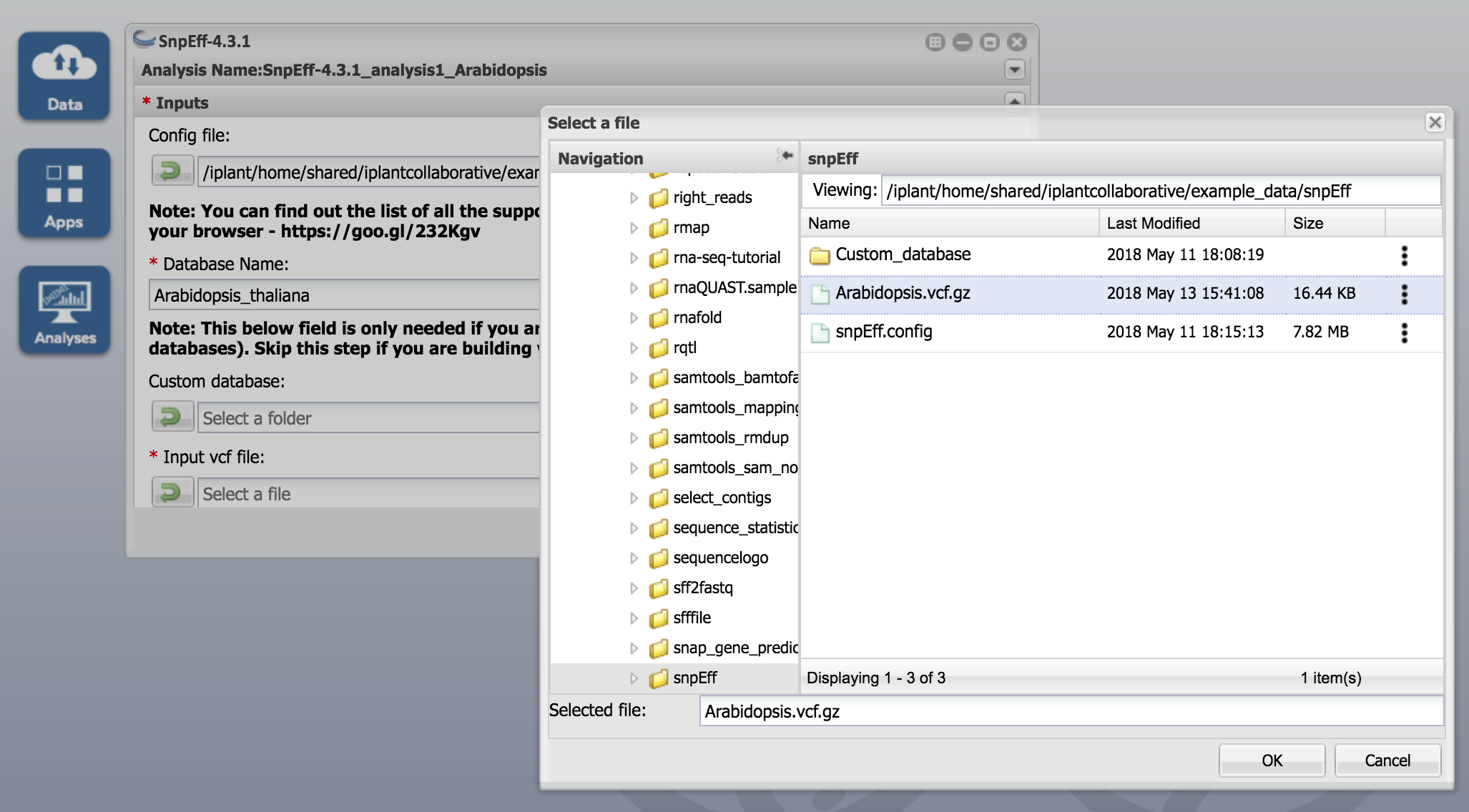
- Use snpEff.config for Config file
- Use Arabidopsis_thaliana for Database Name
- Skip Custom database option. This is for building custom database only
- Use Arabidopsis.vcf.gz for Input vcf file argument
Output:
- Use Arabidopsis_annotated.vcf for Output File Name and then click Launch Analysis
After a successful run you should the output file Arabidopsis_annotated.vcf that contains annotations of the vcf file. Here are the first few lines of the annotated vcf file.
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT 88 1 38751 . AT A 40.0 PASS DP=21687;AC=0;AN=0;ANN=A|splice_region_variant|LOW|AT1G01070|AT1G01070|transcript|AT1G01070.1|protein_coding|6/6|c.*146delA||||||,A|splice_region_variant|LOW|AT1G01070|AT1G01070|transcript|AT1G01070.2|protein_coding|5/5|c.*146delA||||||,A|3_prime_UTR_variant|MODIFIER|AT1G01070|AT1G01070|transcript|AT1G01070.1|protein_coding|6/6|c.*146delA|||||146|,A|3_prime_UTR_variant|MODIFIER|AT1G01070|AT1G01070|transcript|AT1G01070.2|protein_coding|5/5|c.*146delA|||||146|,A|upstream_gene_variant|MODIFIER|LHY|AT1G01060|transcript|AT1G01060.1|protein_coding||c.-1691delA|||||912|,A|upstream_gene_variant|MODIFIER|LHY|AT1G01060|transcript|AT1G01060.3|protein_coding||c.-1691delA|||||995|,A|upstream_gene_variant|MODIFIER|LHY|AT1G01060|transcript|AT1G01060.2|protein_coding||c.-1691delA|||||972|,A|upstream_gene_variant|MODIFIER|LHY|AT1G01060|transcript|AT1G01060.4|protein_coding||c.-1691delA|||||972|,A|upstream_gene_variant|MODIFIER|LHY|AT1G01060|transcript|AT1G01060.5|protein_coding||c.-1916delA|||||881| GT:GQ:DP ./.:38:8 1 38752 . T . 40.0 PASS DP=21532;AN=2 GT:GQ:DP 0|0:38:8 1 38753 . A . 40.0 PASS DP=21709;AN=2 GT:GQ:DP 0|0:38:8 1 38754 . A . 40.0 PASS DP=21296;AN=2 GT:GQ:DP 0|0:38:8 1 38755 . C . 40.0 PASS DP=20865;AN=2 GT:GQ:DP 0|0:36:6 1 38756 . C . 40.0 PASS DP=20817;AN=2 GT:GQ:DP 0|0:36:6 1 38757 . A . 40.0 PASS DP=21015;AN=2 GT:GQ:DP 0|0:36:6 1 38758 . A . 40.0 PASS DP=20973;AN=2 GT:GQ:DP 0|0:36:5 1 38759 . A . 40.0 PASS DP=20831;AN=2 GT:GQ:DP 0|0:38:5 1 38760 . C . 40.0 PASS DP=21080;AN=2 GT:GQ:DP 0|0:38:5 1 38761 . A . 40.0 PASS DP=21130;AN=2 GT:GQ:DP 0|0:38:5 1 38762 . A . 40.0 PASS DP=21252;AN=2 GT:GQ:DP 0|0:38:5 1 38763 . A . 40.0 PASS DP=21062;AN=2 GT:GQ:DP 0|0:38:5 1 38764 . C . 40.0 PASS DP=21284;AN=2 GT:GQ:DP 0|0:36:5 1 38765 . C . 40.0 PASS DP=21283;AN=2 GT:GQ:DP 0|0:36:6 1 38766 . G . 40.0 PASS DP=21247;AN=2 GT:GQ:DP 0|0:36:6 1 38767 . G . 40.0 PASS DP=21332;AN=0 GT:GQ:DP ./.:.:. 1 38768 . A . 40.0 PASS DP=21398;AN=2 GT:GQ:DP 0|0:36:6 1 38769 . T . 40.0 PASS DP=21538;AN=2 GT:GQ:DP 0|0:38:6 1 38770 . T . 40.0 PASS DP=21469;AN=2 GT:GQ:DP 0|0:38:6
As you can see, SnpEff added functional annotations in the ANN info field (eighth column in the VCF output file). Details about the 'ANN' field format can be found in the ANN Field section.
Older SnpEff version used 'EFF' field (details about the 'EFF' field format can be found in the EFF Field section).
2.2 Build your own SnpEff database
Most people do NOT need to build a database, and can safely use a pre-built one. So unless you are working with a rare genome you most likely don't need to do it either.
You need to create our own config-file for SnpEff if your genome is not in the list of the database in this file snpeff_databases.csv. If your genome has already had a database, you can skip to running SnpEff step
In order to build a database for a new genome, you need to:
- Configure a new genome in SnpEff's config file
snpEff.config.- Add genome entry to snpEff's configuration
- If the genome uses a non-standard codon table: Add codon table parameter
- Get the reference genome sequence (e.g. in FASTA format).
- Get genome annotations. There are four different ways you can do this:
All files can be compressed using gzip. E.g. the reference file 'hg19.fa' can be compressed to 'hg19.fa.gz', snpEff will automatically decompress the file.
Some files claimed to be compressed using GZIP are actually not or even use a block compression variant not supported by Java's gzip library. If you notice that your build process finishes abruptly for no apparent reason, try uncompressing the files. This sometimes happens with ENSEMBL files.
Let's suppose you are working on a genome that is not currently supported, then the following example applies. Here I am trying to build a database for a version of Brassica rapa that is currently not supported in SnpEff data.2.2.1 Build data first
All files are located for Basic example is located in the Community Data directory of the CyVerse Discovery Environment at the following path:
Community Data > iplantcollaborative > example_data > snpEff > Custom_database (/iplant/home/shared/iplantcollaborative/example_data/snpEff/Custom_database)
2.2.1.1 After logging into Discovery Environment, click on the app window and in the search box, enter snpeff. You will see two apps - SnpEff-4.3.1 and SnpEff-build-4.3.1
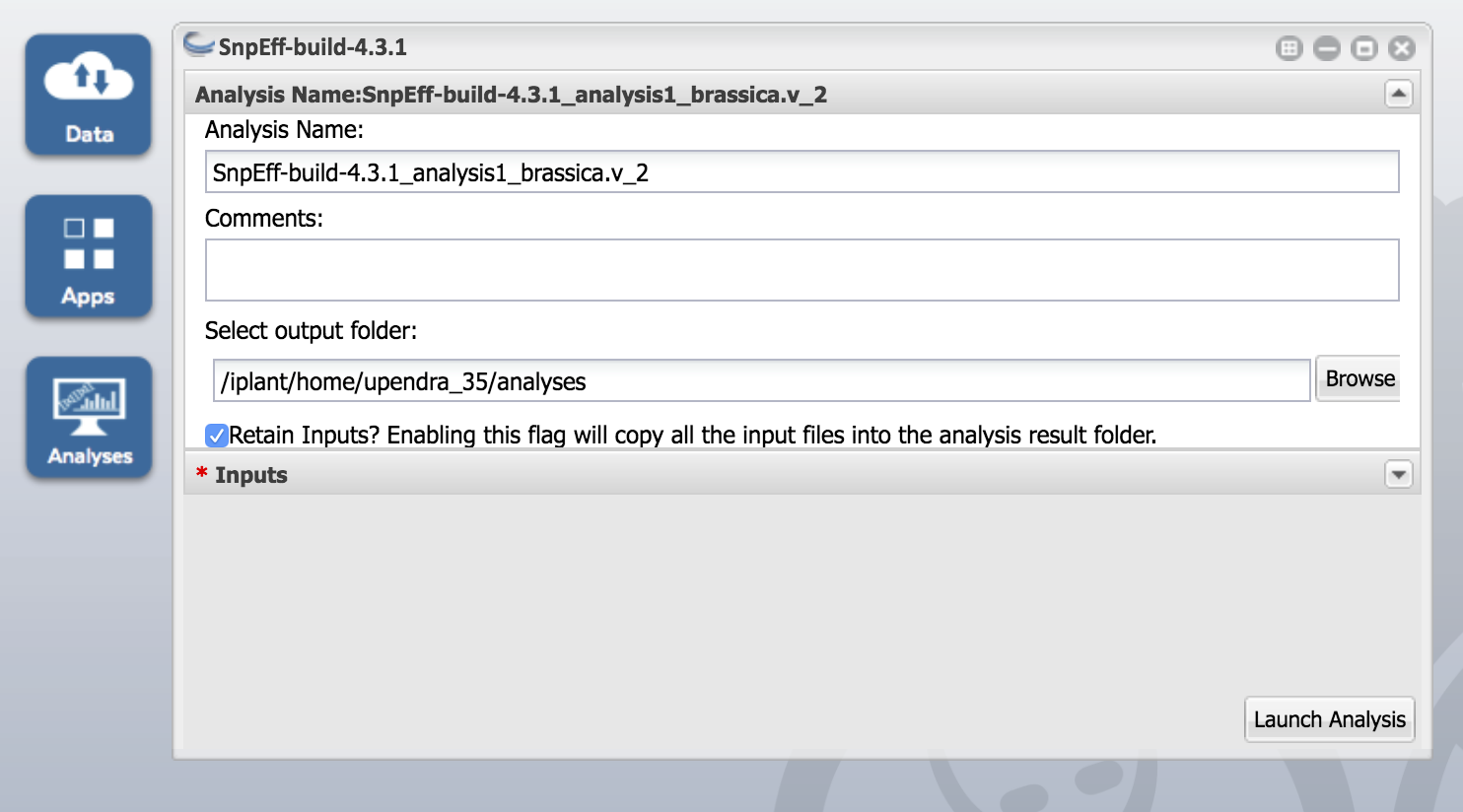
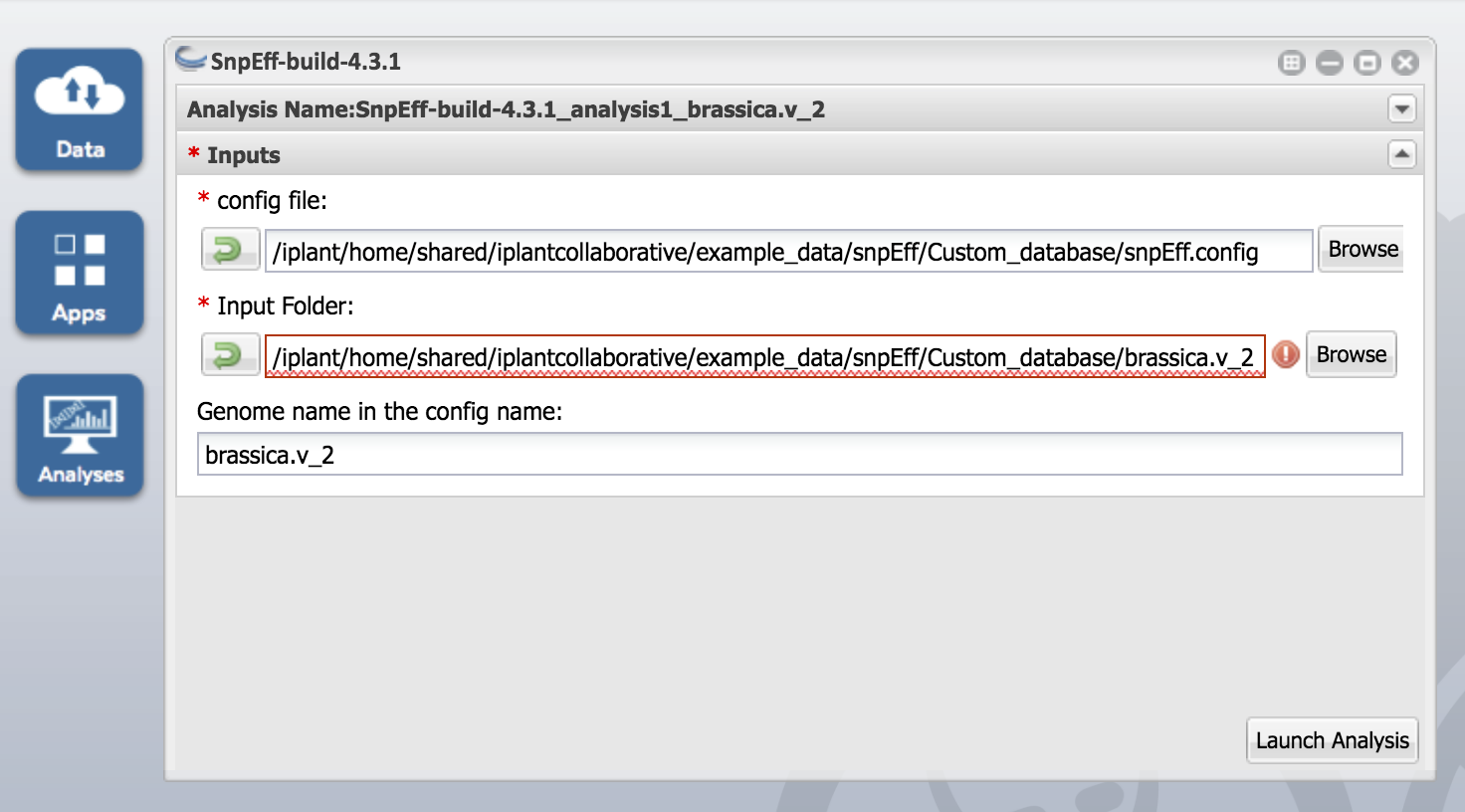
Click on the SnpEff-build-4.3.1 app and enter "SnpEff-4.3.1_analysis1_brassica.v_2" under the Analysis Name.
Make sure you check the box that says "Retain Inputs? Enabling this flag will copy all the input files into the analysis results folder." Otherwise the build doesn't work
Inputs:
- Use snpEff.config as input for config file
In order to tell SnpEff that there is a new genome available, you must update SnpEff's configuration file snpEff.config. You must add a new genome entry to snpEff.config.
If your genome, or a chromosome, uses non-standard codon tables you must update snpEff.config accordingly. A typical case is when you use mitochondrial DNA. Then you specify that chromosome 'MT' uses codon.Invertebrate_Mitochondrial codon table. Another common case is when you are adding a bacterial genome, then you specify that the codon table is Bacterial_and_Plant_Plastid.
This config file snpEff.config has the custom genome (brassica_v.2) added to the configuration file. If you want to add your own custom genome, then you can download this snpEff.config file and edit this file to add the following two lines after #Non-standard Database section. Here is an example
#---
# Non-standard Databases
#---
# My Brassica genome
brassica.v_2.genome = BrassicaRapa
2. Use brassica.v_2 as Input folder
This config file snpEff.config has the custom genome (brassica_v.2) added to the configuration file. If you want to add your own custom genome, then you can download this snpEff.config file and put your genome after #Non-standard Database section. Here is an example
#---
# Non-standard Databases
#---
# My Brassica genome
brassica.v_2.genome = BrassicaRapa
What are the contents in brassica.v_2 folder? If you look inside this folder, you will find only two files. Compressed genome fasta file and renamed as sequences.fa.gz and compressed genome annotation gff3 file renamed as genes.gff.gz. In addition, the folder name brassica.v_2 should match the first of the name in the config file. For example brassica.v_2.genome
3. Finally Genome name in the config name should be brassica.v_2. Again make sure that this name should match the names in the config file and input folder. Then click Launch Analysis button
After successful completion of the build, you will get three outputs:
- brassica.v_2 folder that contains sequences.fa.gz, genes.gff.gz and snpEffectPredictor.bin files
- logs folder
- snpEff.config file
We need brassica.v_2 and snpEff.config for the next step
2.2.1.2 Run snpeff using the custom build
- Click on the SnpEff-4.3.1 app and enter "SnpEff-4.3.1_analysis1_brassica" under the Analysis Name
- Inputs:
- Use snpEff.config Config file from above step
- Use brassica.v_2 for Database Name
- Use brassica.v_2 folder from above step
- Use Brassica_rapa.vcf.gz as input vcf file
- Outputs:
- Use brassica_annotated.vcf as output vcf file
After successful completion of snpeff analysis, you should get the brassica_annotated.vcf that contains the annotated vcfs for our custom database (not present in snpeff's database)